Principal Investigator
- Andrew F. Laine, D.Sc., HBIL
- Graham Barr, MD-PhD, Department of Medicine, Columbia University Medical Center
Students/Research Scientists
- Elsa Angelini, PhD.
- Jie Yang BSc
Collaborators
- John Austin M.D., Department of Radiology, Columbia University Medical Center
- Pallavi Balte MSc, Department of Medicine, Columbia University Medical Center
- Eric Hoffman MD, Department of Radiology, Iowa University
- Daniel Rabinowitz PHD, Department of Statistics, Columbia University
Summary
Pulmonary emphysema is a condition involving alveolar wall destruction, contributing to chronic airflow limitation characteristic of chronic obstructive pulmonary disease (COPD). The processes underlying COPD are currently not well understood, even though the disease is a leading cause of morbidity and mortality worldwide. Computed tomography (CT) is a routinely performed imaging study to identify and quantify the extent of emphysema. A single CT volume holds a tremendous amount of data, but the current lack of dedicated image analysis methods restricts the amount and quality of information that can be extracted from the images. Our research is focused on two main topics:
1) Adaptive segmentation and quantification of emphysema, and
2) Unsupervised learning of Lung Texture Patterns
Adaptive segmentation and quantification of emphysema
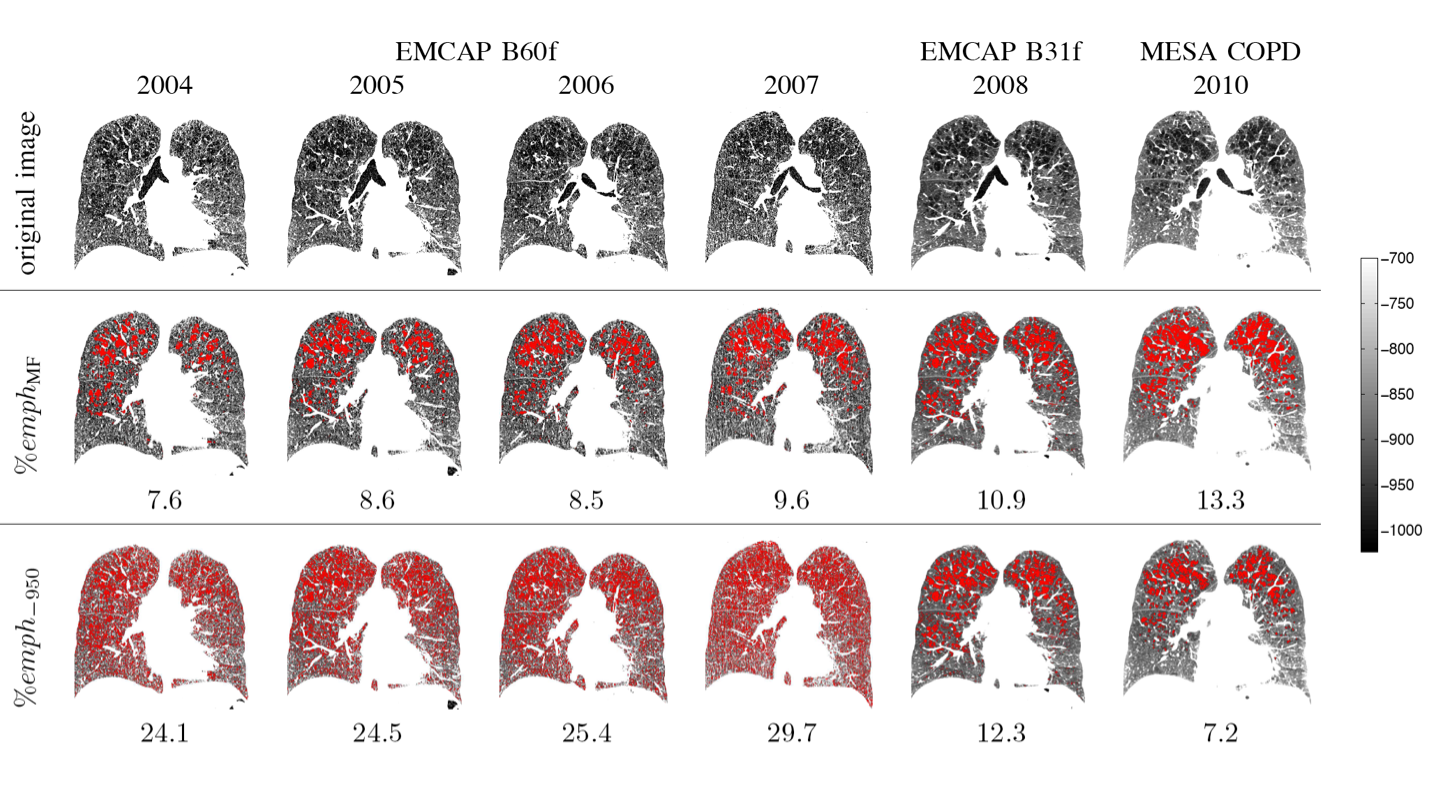
Estimation of emphysema extent is generally done by quantifying the relative lung area below a fixed attenuation threshold. However, due to the variability in intensity distributions, emphysema measures are usually not comparable between imaging studies with varying imaging protocols. Our work uses a Hidden Markov field models to provide a more reliable measure of the condition.
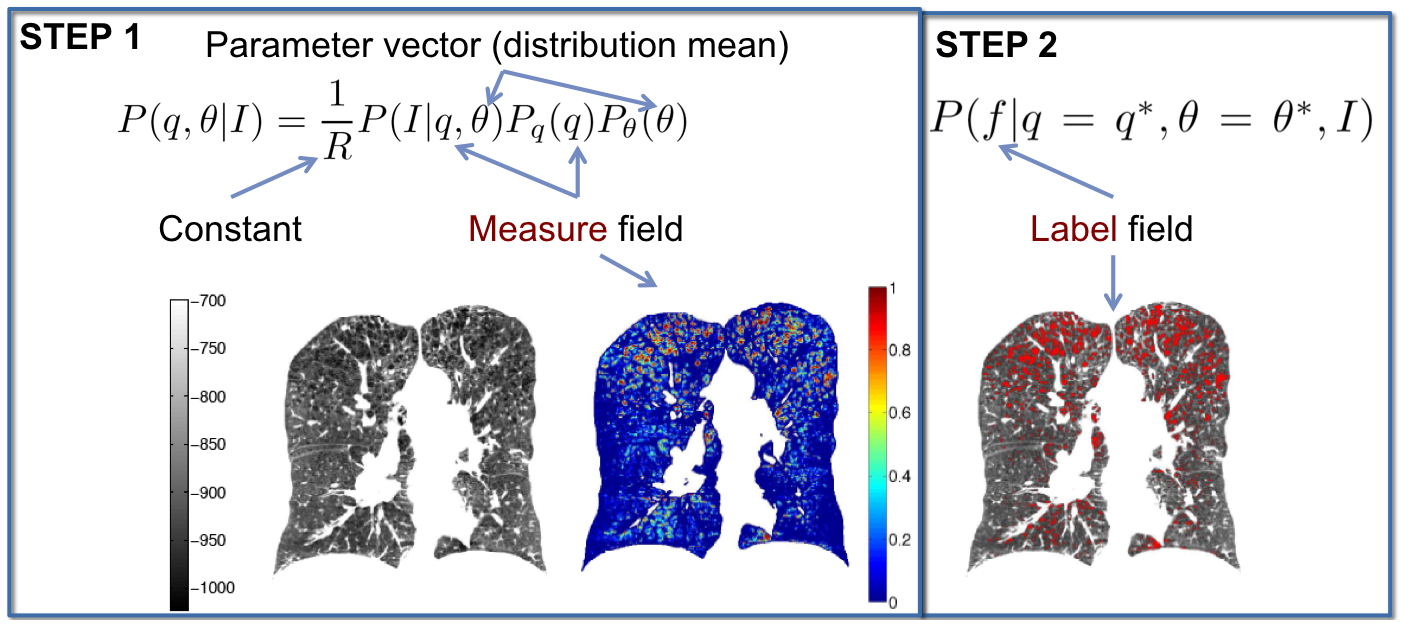
The results of this work have several applications, especially for longitudinal studies and patient-specific assessment of emphysema.
Unsupervised learning of Lung Texture Patterns (LTPs)
Current use of textural image information for subtyping emphysema is limited to supervised learning, used to replicate manual labeling. Our work is aimed at unsupervised methods to leverage large image datasets to learn lung tissue representations.
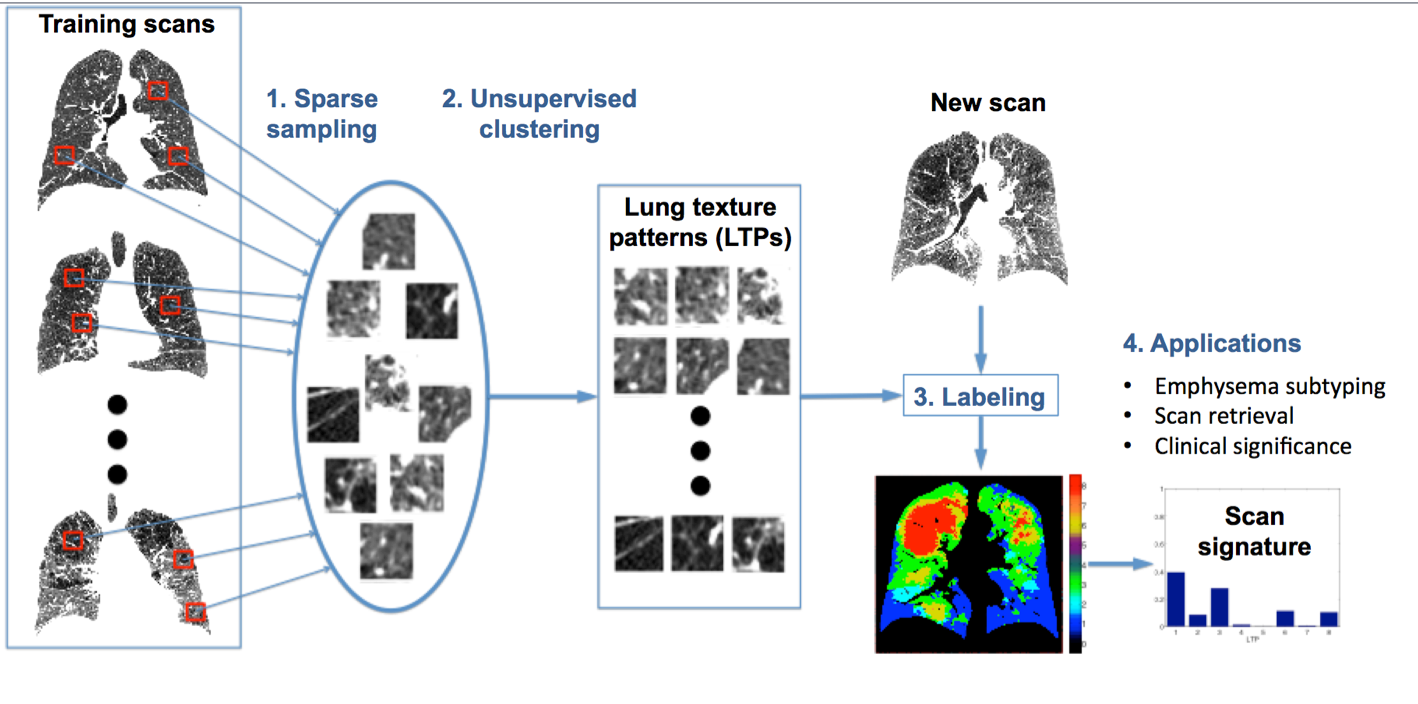
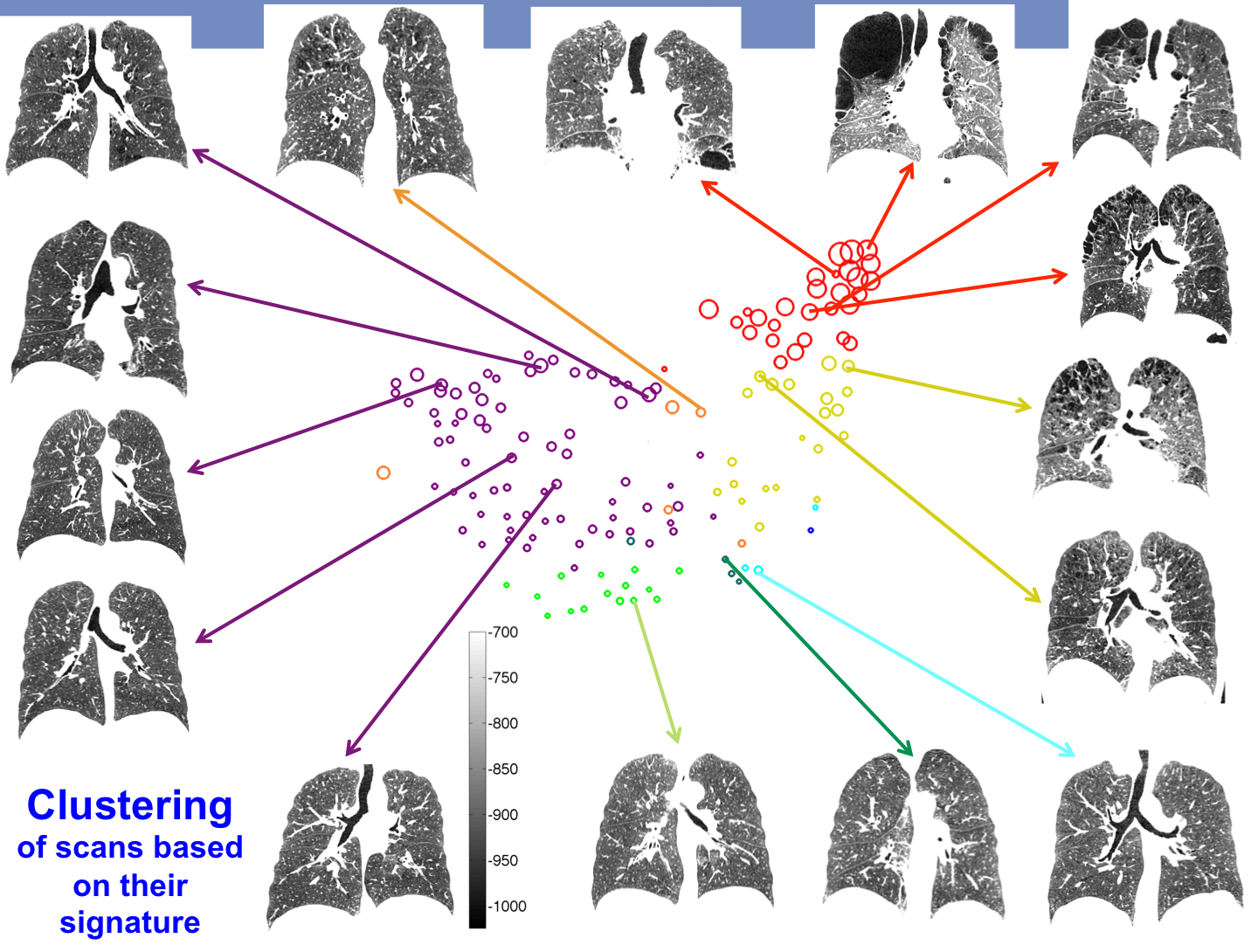
The learned LTPs can be used for example for patient clustering using their LTP histograms as their signature.
Publications
Yrjö Häme, Elsa D. Angelini, Eric A. Hoffman, R. Graham Barr, Andrew F Laine: “Robust quantification of pulmonary emphysema with a hidden Markov measure field model”, IEEE International Symposium on Biomedical Imaging (ISBI), San Francisco, CA, USA, 2013
Y. Hame, E. Angelini, M. Patrick, B. Smith, E. Hoffman, G. R. Barr, A. F. Laine, "Equating
Emphysema Scores and Segmentations across CT Reconstructions: A Comparison Study",
IEEE International Symposium on Biomedical Imaging (ISBI), Brooklyn, NY, USA, 2015.
Y. Hame, E. Angelini, E. Hoffman, G. Barr, A. Laine, "Adaptive quantification and longitudinal analysis of pulmonary emphysema with a hidden Markov measure field model", IEEE Transactions on Medical Imaging, Vol. 33, No.7, pp. 1527 - 1540, 2014.
Y. Hame, E. Angelini, M. Patrick, B. Smith, E. Hoffman, G. R. Barr, A. F. Laine, "Sparse Sampling and Unsupervised Learning of Lung Texture Patterns in Pulmonary Emphysema: MESA COPD Study", IEEE International Symposium on Biomedical Imaging (ISBI), Brooklyn,
NY, USA, 2015.
